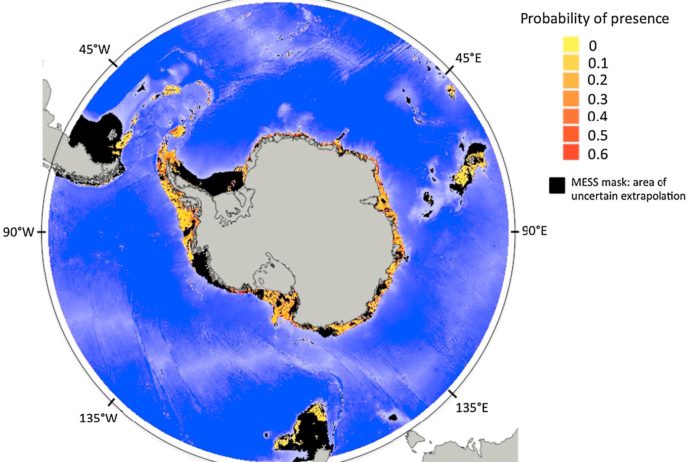
A new research paper by Charlène Guillaumot et al. “Broad-scale species distribution models applied to data-poor areas” has just been published in Progress in Oceanography. The paper specifically addresses Species Distribution Models, as they have been increasingly used over the past decades to characterise the spatial distribution and the ecological niche of various taxa. Validating predicted species distribution is important, especially when producing broad-scale models (i.e. at continental or oceanic scale) based on limited and spatially aggregated presence-only records. In the present study, several model calibration methods are compared and guidelines are provided to perform relevant SDMs using a Southern Ocean marine species, the starfish Odontaster validus Koehler, 1906, as a case study. The effect of the spatial aggregation of presence-only records on modelling performance is evaluated and the relevance of a target-background sampling procedure to correct for this effect is assessed. The accuracy of model validation is estimated using k-fold random and spatial cross- validation procedures. Finally, we evaluate the relevance of the Multivariate Environmental Similarity Surface (MESS) index to identify areas in which SDMs accurately interpolate and conversely, areas in which models extrapolate outside the environmental range of occurrence records. Results show that the random cross-validation procedure (i.e. a widely applied method, for which training
and test records are randomly selected in space) tends to over-estimate model performance when applied to spatially aggregated datasets. Spatial cross-validation procedures can compensate for this over-estimation effect but different spatial cross-validation procedures must be tested for their ability to reduce over-fitting while providing relevant validation scores. Model predictions show that SDM generalisation is limited when working with aggregated datasets at broad spatial scale. The MESS index calculated in our case study show that over half of the predicted area is highly uncertain due to extrapolation. Our work provides methodological guidelines to generate accurate model assessments at broad spatial scale when using limited and aggregated presence-only datasets. We highlight the importance of taking into account the presence of spatial aggregation in species records and using non-random cross-validation procedures. Evaluating the best calibration procedures and correcting for spatial biases should be considered ahead the modelling exercise to improve modelling relevance.
The full paper can be found here.